A team of researchers from China’s Shanghai Jiao Tong University have created what may be the world’s first programmable DNA computer.
In a paper published in prestigious magazine Nature they explained how they created DNA-based programmable gate arrays or “DPGAs” that can support more than 100 billion distinct computational circuits.
Their success represents what they described as “a key step towards general-purpose DNA computing” and allowed them to classify molecules and solve quadratic equations using the “biocomputer.”
This, admittedly, did not happen fast: silicon is not yet at risk...
DNA, you say?
The idea of using DNA for computational purposes is not new.
Microsoft has been among the organisations exploring it for storage and compute – successfully storing data on synthetic DNA and in 2017 demonstrating a way of creating DNA logic gates and interconnects.
Researchers at Harvard in 2017 meanwhile built a biological computer inside living E. coli cells; successfully using it to store a 36-by-26-pixel GIF of one of the first moving images ever recorded.
The “biocomputing” approach relies on exploiting biomolecular interactions to work – yet most systems to date have only been able to implement a specific algorithm, or limited number of computing tasks.
The recent paper, showing success at developing programmable arrays of logic gates, suggests a genuinely major breakthrough in the field.
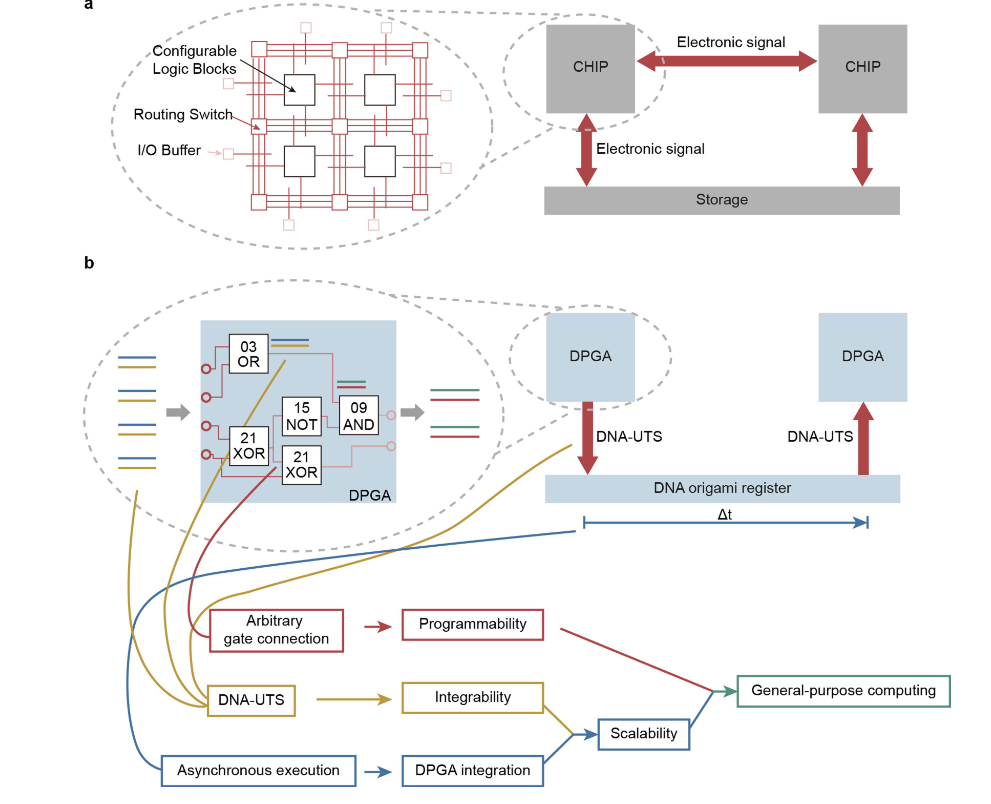
As its authors put it: “We find that the use of generic single-stranded oligonucleotides as a uniform transmission signal can reliably integrate large-scale DICs (DNA integrated circuits) with minimal leakage and high fidelity for general-purpose computing” – with a single DPGA able to be programmed to implement over 100 billion distinct circuits.
DNA computing: China’s new success
DNA consists of strands made up of four different molecules or “bases”: adenine, thymine, cytosine, and guanine. abbreviated as ATCG. In DNA computing, these can be used to encode the binary number pairs 00, 01, 10, and 11 – then help deliver computations that are based on the specific way in which these bases bind to each other.
Specific sequences of DNA molecules can then be designed to bind and come apart in a way that lets them function as logic gates; which, crudely, accept inputs and then output results based on their state.
The team of researchers from Shanghai used this approach to create a quadratic equation-solving DNA computer (DIC) “assembled with three layers of cascade DPGAs comprising 30 logic gates with around 500 DNA strands. We further show that integration of a DPGA with an analog-to-digital converter can classify disease-related microRNAs.”
As Charles Q. Choi, writing for IEEE Spectrum notes: “A key obstacle standing in the way of DNA computing is that DNA molecules can flow in essentially any direction. This makes it challenging to bring logic gates together to perform computations in programmed sequences.
“To surmount this problem, the researchers built so-called DNA origami. By designing a DNA sequence just right, one can get the resulting floppy strand to stick to itself and bend into virtually whatever 2D or 3D shape is desired. The DNA origami can fold and hold together because each DNA base binds to a specific partner. The scientists generated DNA origami that could act like registers—devices that guide the flow of data and instructions within computers. These helped control the intrinsically random collisions of DNA molecules.”
The DNA computer, given 18 diseased and five healthy genetic samples, correctly detected and reported which were which – in about 2 hours.
Such computers could be “useful in biomedical applications—for example, cellular programming and molecular diagnostics,” study co-author Fei Wang told Choi, who notes that “since DNA computers use DNA as both input and output material, one can design them to, say, respond to a gene they detect by releasing a strand of DNA that can in turn have biological effects” and “may find use in programming cells to respond to… cancer-related molecules for disease treatment.”
Tempted to try this at home?
Preparation of dual-rail gates and wiring instructions involved dissolving DNA oligonucleotides in: “1× TE buffer (nuclease free,pH 8.0, Sigma-Aldrich), quantified with ultraviolet/visible spectrometry by monitoring their absorption at 260 nm and storage at −20 °C. Oligonucleotides labelled with fluorescent dyes or quenchers were dissolved in deionized water… and stored in deionized water at −20 °C. Single strands were hybridized to prepare corresponding double-stranded DNA… Hybridized structures were prepared by mixing the required strands to the corresponding final concentrations in TE buffer (1× TE: 40 mM Tris base, 20 mM acetic acid, 2 mM EDTA adjusted to pH 8.0) with 12.5 mM MgCl.”
Upgrading that RAM may be a little easier.
The source codes used in the DNA computing study (Visual DSD, MATLAB, Python) are available from GitHub here.





